Publications
2025 / Direct RNA Oxford Nanopore sequencing distinguishes between modifications in tRNA at the U position
W. Kusmirek, N. Strozynska, . Martin-Arroyo Cerpa, A. Dziergowska, G. Leszczynska, R. Nowak, M. Adamczyk / International Journal of Biological Macromolecules, 2025, vol. 320, s.1-14. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.1016/j.ijbiomac.2025.145877
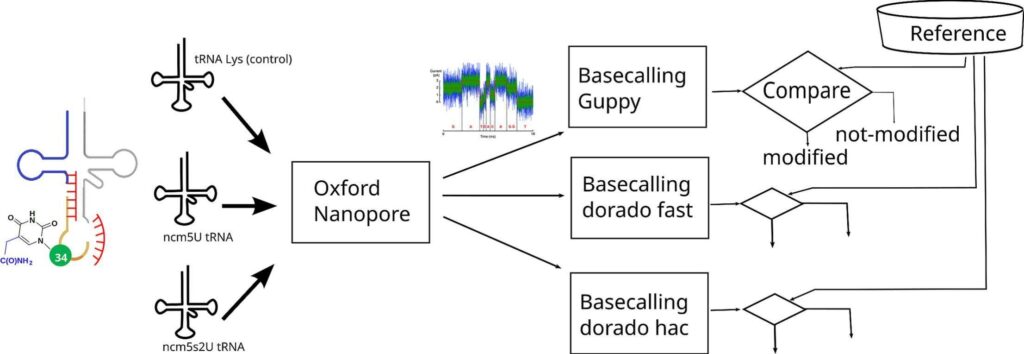
The measurement of tRNA modifications with single transcript resolution has been feasible for only a few modifications due to the lack of available methods. This limitation doesn’t allow to advance studies on the dynamic nature of tRNA modification and its cellular function in time and space, neither to develop modern diagnostic tools for several already known tRNA-dependent human diseases. Oxford Nanopore Sequencing (ONS) is a method that has proven to be efficient for the study of mRNA. The analysis of tRNA modifications by ONS is still under development. We have developed new methods to synthesise modified tRNA macromolecules with xcm5U and xcm5s2U modifications at the anticodon loop. We have investigated the efficacy of ONS to discriminate between complex modifications of uridine 34 in singly modified tRNA, that are difficult to accurately predict. ONS captures the features produced by uridine with a thiol group (s2U) and without thiol when present on synthetic tRNA. Thus, ONS has a great potential for developing strategies to accurately identify the modification status of the tRNA anticodon loop, which encompasses the most complex modifications on uridine-containing RNA motifs. Thio-modification of U34 in tRNA is associated with a group of deadliest diseases.
2024 / Mitochondrial Aconitase and Its Contribution to the Pathogenesis of Neurodegenerative Diseases
Padalko Volodymyr, Pośnik Filip, Adamczyk Małgorzata / International Journal of Molecular Sciences, 2024, vol. 25, nr 18, s.1-33, Numer artykułu:9950. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.3390/ijms25189950
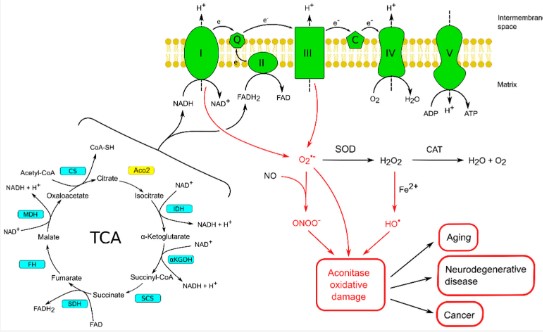
This survey reviews modern ideas on the structure and functions of mitochondrial and cytosolic aconitase isoenzymes in eukaryotes. Cumulative experimental evidence about mitochondrial aconitases (Aco2) as one of the main targets of reactive oxygen and nitrogen species is generalized. The important role of Aco2 in maintenance of homeostasis of the intracellular iron pool and maintenance of the mitochondrial DNA is discussed. The role of Aco2 in the pathogenesis of some neurodegenerative diseases is highlighted. Inactivation or dysfunction of Aco2 as well as mutations found in the ACO2 gene appear to be significant factors in the development and promotion of various types of neurodegenerative diseases. A restoration of efficient mitochondrial functioning as a source of energy for the cell by targeting Aco2 seems to be one of the promising therapeutic directions to minimize progressive neurodegenerative disorders.
2023 / Mitochondrial Metabolism in the Spotlight: Maintaining Balanced RNAP III Activity Ensures Cellular Homeostasis
Szatkowska Róża, Emil Furmanek, Andrzej M. Kierzak, Ludwig Christian, Małgorzata Adamczyk / International Journal of Molecular Sciences, 2023, vol. 24, nr 19, s.1-24, Numer artykułu:14763. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.3390/ijms241914763

RNA polymerase III (RNAP III) holoenzyme activity and the processing of its products have been linked to several metabolic dysfunctions in lower and higher eukaryotes. Alterations in the activity of RNAP III-driven synthesis of non-coding RNA cause extensive changes in glucose metabolism. Increased RNAP III activity in the S. cerevisiae maf1Δ strain is lethal when grown on a non-fermentable carbon source. This lethal phenotype is suppressed by reducing tRNA synthesis. Neither the cause of the lack of growth nor the underlying molecular mechanism have been deciphered, and this area has been awaiting scientific explanation for a decade. Our previous proteomics data suggested mitochondrial dysfunction in the strain. Using model mutant strains maf1Δ (with increased tRNA abundance) and rpc128-1007 (with reduced tRNA abundance), we collected data showing major changes in the TCA cycle metabolism of the mutants that explain the phenotypic observations. Based on 13C flux data and analysis of TCA enzyme activities, the present study identifies the flux constraints in the mitochondrial metabolic network. The lack of growth is associated with a decrease in TCA cycle activity and downregulation of the flux towards glutamate, aspartate and phosphoenolpyruvate (PEP), the metabolic intermediate feeding the gluconeogenic pathway. rpc128-1007, the strain that is unable to increase tRNA synthesis due to a mutation in the C128 subunit, has increased TCA cycle activity under non-fermentable conditions. To summarize, cells with non-optimal activity of RNAP III undergo substantial adaptation to a new metabolic state, which makes them vulnerable under specific growth conditions. Our results strongly suggest that balanced, non-coding RNA synthesis that is coupled to glucose signaling is a fundamental requirement to sustain a cell’s intracellular homeostasis and flexibility under changing growth conditions. The presented results provide insight into the possible role of RNAP III in the mitochondrial metabolism of other cell types.
2022 / Making Sense of “Nonsense” and More: Challenges and Opportunities in the Genetic Code Expansion, in the World of tRNA Modifications
Lateef Olubodun Michael, Akintubosun Michael Olawale, Olaoba Olamide Tosin, Sunday Ocholi Samson, Małgorzata Adamczyk / International Journal of Molecular Sciences, 2022, vol. 23, nr 2, s.1-33, Numer artykułu:938. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.3390/ijms23020938
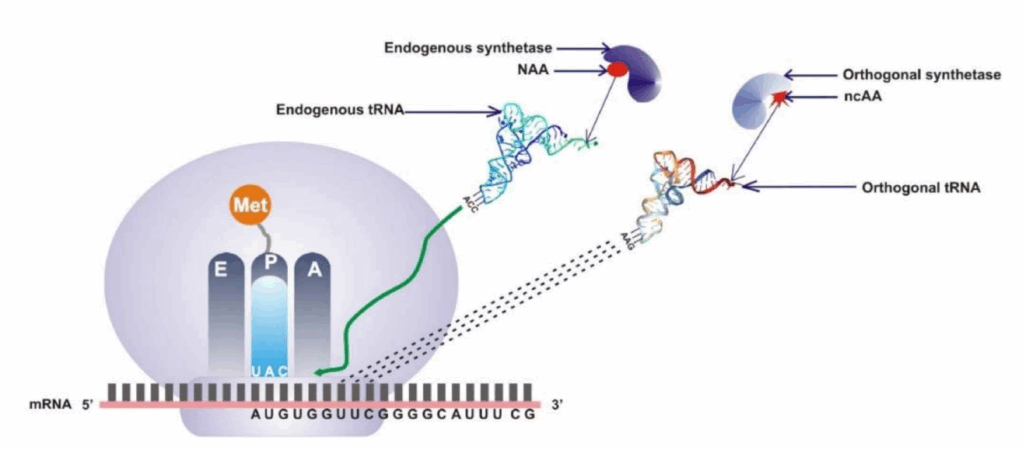
The evolutional development of the RNA translation process that leads to protein synthesis based on naturally occurring amino acids has its continuation via synthetic biology, the so-called rational bioengineering. Genetic code expansion (GCE) explores beyond the natural translational processes to further enhance the structural properties and augment the functionality of a wide range of proteins. Prokaryotic and eukaryotic ribosomal machinery have been proven to accept engineered tRNAs from orthogonal organisms to efficiently incorporate noncanonical amino acids (ncAAs) with rationally designed side chains. These side chains can be reactive or functional groups, which can be extensively utilized in biochemical, biophysical, and cellular studies. Genetic code extension offers the contingency of introducing more than one ncAA into protein through frameshift suppression, multi-site-specific incorporation of ncAAs, thereby increasing the vast number of possible applications. However, different mediating factors reduce the yield and efficiency of ncAA incorporation into synthetic proteins. In this review, we comment on the recent advancements in genetic code expansion to signify the relevance of systems biology in improving ncAA incorporation efficiency. We discuss the emerging impact of tRNA modifications and metabolism in protein design. We also provide examples of the latest successful accomplishments in synthetic protein therapeutics and show how codon expansion has been employed in various scientific and biotechnological applications.
2021 / Unique Properties of the Alpha-Helical DNA-Binding Protein KfrA Encoded by the IncU Incompatibility Group Plasmid RA3 and Its Host-Dependent Role in Plasmid Maintenance
Lewicka Ewa, Mitura Monika, Steczkiewicz Kamil, Justyna Kieracinska, Kamila Skrzynska, Małgorzata Adamczyk, Grażyna Jagura-Burdzy / Applied and Environmental Microbiology, 2021, vol. 87, nr 2, s.1-27, Numer artykułu:e01771-20. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.1128/aem.01771-20

Alpha-helical coiled-coil KfrA-type proteins are encoded by various broad-host-range low-copy-number conjugative plasmids. The DNA-binding protein KfrA encoded on the RA3 plasmid, a member of the IncU incompatibility group, oligomerizes, forms a complex with another plasmid-encoded, alpha-helical protein, KfrC, and interacts with the segrosome proteins IncC and KorB. The unique mode of KfrA dimer binding to the repetitive operator is required for a KfrA role in the stable maintenance of RA3 plasmid in distinct hosts.
2021 / Revealing biophysical properties of KfrA-type proteins as a novel class of cytoskeletal, coiled-coil plasmid-encoded proteins
Małgorzata Adamczyk, Ewa Lewicka, Róża Szatkowska, H. Nieznanska, J. Ludwiczak, M. Jasiński, Stanislaw Dunin-Horkawicz, E. Sitkiewicz, B. Swiderska, G. Goch, Grażyna Jagura-Burdzy / BMC Microbiology, 2021, vol. 21, nr 1, s.1-16, Numer artykułu:32. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.1186/s12866-020-02079-w

AbstractBackgroundDNA binding KfrA-type proteins of broad-host-range bacterial plasmids belonging to IncP-1 and IncU incompatibility groups are characterized by globular N-terminal head domains and long alpha-helical coiled-coil tails. They have been shown to act as transcriptional auto-regulators.ResultsThis study was focused on two members of the growing family of KfrA-type proteins encoded by the broad-host-range plasmids, R751 of IncP-1β and RA3 of IncU groups. Comparative in vitro and in silico studies on KfrAR751and KfrARA3confirmed their similar biophysical properties despite low conservation of the amino acid sequences. They form a wide range of oligomeric forms in vitro and, in the presence of their cognate DNA binding sites, they polymerize into the higher order filaments visualized as “threads” by negative staining electron microscopy. The studies revealed also temperature-dependent changes in the coiled-coil segment of KfrA proteins that is involved in the stabilization of dimers required for DNA interactions.ConclusionKfrAR751and KfrARA3are structural homologues. We postulate that KfrA type proteins have moonlighting activity. They not only act as transcriptional auto-regulators but form cytoskeletal structures, which might facilitate plasmid DNA delivery and positioning in the cells before cell division, involving thermal energy.
2021 / Alpha-Helical Protein KfrC Acts as a Switch between the Lateral and Vertical Modes of Dissemination of Broad-Host-Range RA3 Plasmid from IncU (IncP-6) Incompatibility Group
Monika Mitura, Ewa Lewicka, Jolanta Godziszewska, Małgorzata Adamczyk, Grażyna Jagura-Burdzy / International Journal of Molecular Sciences, 2021, vol. 22, nr 9, s.1-34, Numer artykułu:4880. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.3390/ijms22094880
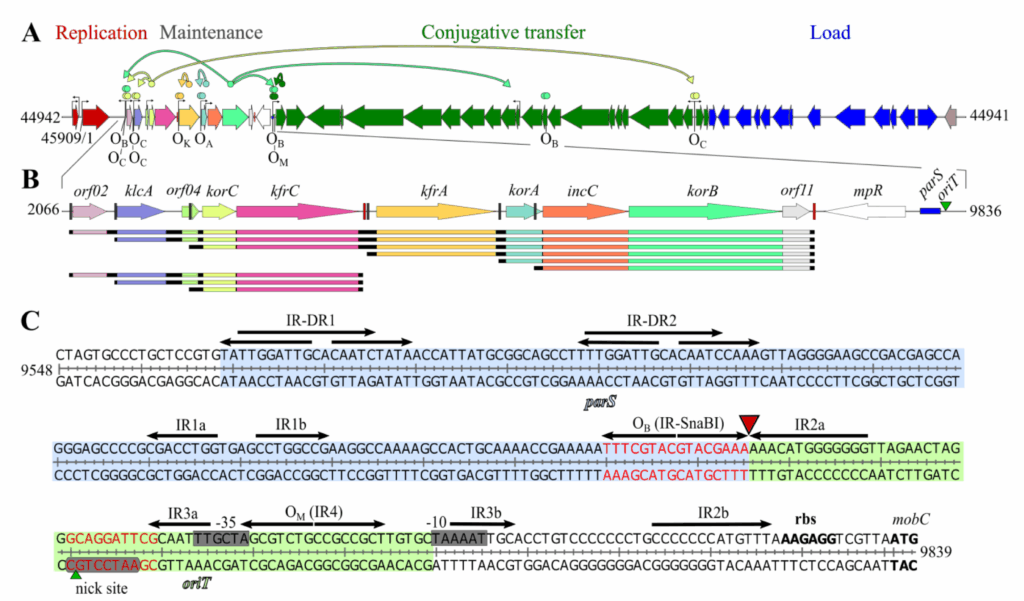
KfrC proteins are encoded by the conjugative broad-host-range plasmids that also encode alpha-helical filament-forming KfrA proteins as exemplified by the RA3 plasmid from the IncU incompatibility group. The RA3 variants impaired in kfrA, kfrC, or both affected the host’s growth and demonstrated the altered stability in a species-specific manner. In a search for partners of the alpha-helical KfrC protein, the host’s membrane proteins and four RA3-encoded proteins were found, including the filamentous KfrA protein, segrosome protein KorB, and the T4SS proteins, the coupling protein VirD4 and ATPase VirB4. The C-terminal, 112-residue dimerization domain of KfrC was involved in the interactions with KorB, the master player of the active partition, and VirD4, a key component of the conjugative transfer process. In Pseudomonas putida, but not in Escherichia coli, the lack of KfrC decreased the stability but improved the transfer ability. We showed that KfrC and KfrA were involved in the plasmid maintenance and conjugative transfer and that KfrC may play a species-dependent role of a switch between vertical and horizontal modes of RA3 spreading.
2019 / Glycolytic flux in Saccharomyces cerevisiae is dependent on RNA polymerase III and its negative regulator Maf1
Róża Szatkowska, Manuel Garcia-Albornoz, Katarzyna Roszkowska, Stephen W. Holman, Emil Furmanek, Simon J. Hubbard, Robert J. Beynon, Małgorzata Adamczyk / Biochemical Journal, 2019, vol. 476, nr 7, s.1053-1082. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.1042/BCJ20180701

AbstractProtein biosynthesis is energetically costly, is tightly regulated and is coupled to stress conditions including glucose deprivation. RNA polymerase III (RNAP III)-driven transcription of tDNA genes for production of tRNAs is a key element in efficient protein biosynthesis. Here we present an analysis of the effects of altered RNAP III activity on the Saccharomyces cerevisiae proteome and metabolism under glucose-rich conditions. We show for the first time that RNAP III is tightly coupled to the glycolytic system at the molecular systems level. Decreased RNAP III activity or the absence of the RNAP III negative regulator, Maf1 elicit broad changes in the abundance profiles of enzymes engaged in fundamental metabolism in S. cerevisiae. In a mutant compromised in RNAP III activity, there is a repartitioning towards amino acids synthesis de novo at the expense of glycolytic throughput. Conversely, cells lacking Maf1 protein have greater potential for glycolytic flux.
2017 / Low RNA Polymerase III activity results in up regulation of HXT2 glucose transporter independently of glucose signaling and despite changing environment
Adamczyk Małgorzata, Szatkowska Róża / PLoS ONE, 2017, vol. 12, nr 9, s.1-35, Numer artykułu:e0185516. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.1371/journal.pone.0185516

We demonstrate the first evidence that RNA Polymerase III (RNAP III) activity affects the expression of the glucose transporter HXT2 (RNA Polymerase II dependent—RNAP II) at the level of transcription. Down-regulation of RNAP III activity in an rpc128-1007 mutant results in a significant increase in HXT2 mRNA, which is considered to respond only to low extracellular glucose concentrations. HXT2 expression is induced in the mutant regardless of the growth conditions either at high glucose concentration or in the presence of a non-fermentable carbon source such as glycerol. Using chromatin immunoprecipitation (ChIP), we found an increased association of Rgt1 and Tup1 transcription factors with the highly activated HXT2 promoter in the rpc128-1007 strain. Furthermore, by measuring cellular abundance of Mth1 corepressor, we found that in rpc128-1007, HXT2 gene expression was independent from Snf3/Rgt2-Rgt1 (SRR) signaling. The Snf1 protein kinase complex, which needs to be active for the release from glucose repression, also did not appear perturbed in the mutated strain.
2015 / High density monolayers of plasmid protein on latex particles: experiments and theoretical modeling

Monolayers obtained by adsorption of the plasmid protein KfrA on negatively charged polystyrene latex particles under diffusion-controlled conditions at pH 3.5 were interpreted in terms of the random sequential adsorption (RSA) model. A quantitative agreement of the theoretical results derived from these calculations with experimental data was attained for the ionic strength from 0.15 up to 10 −2 M. This confirmed the adsorption mechanism of KfrA molecules on latex in the form of tetramers up to 10 −2 M. On the other hand, for the ionic strength of 10 −3 M the experimental coverage agreed with theoretical predictions under the assumption that screening of electrostatic interaction is enhanced by the presence of counterions and negatively charged polymer chains stemming from latex particles.
2014 / Fructose bisphosphate aldolase is involved in the control of RNA polymerase III-directed transcription
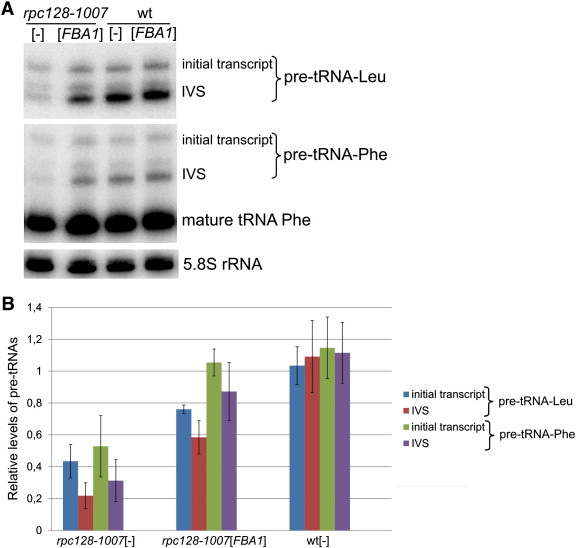
Yeast Fba1 (fructose 1,6-bisphosphate aldolase) is a glycolytic enzyme essential for viability. The overproduction of Fba1 enables the overcoming of a severe growth defect caused by a missense mutation rpc128-1007 in a gene encoding the C128 protein, the second largest subunit of the RNA polymerase III complex. The suppression of the growth phenotype by Fba1 is accompanied by enhanced de novo tRNA transcription in rpc128-1007 cells. We inactivated residues critical for the catalytic activity of Fba1. Overproduction of inactive aldolase still suppressed the rpc128-1007 phenotype, indicating that function of this glycolytic enzyme in RNA polymerase III transcription is independent of its catalytic activity. Yeast Fba1 was determined to interact with the RNA polymerase III complex by coimmunoprecipitation. Additionally, a role of aldolase in control of tRNA transcription was confirmed by ChIP experiments. The results indicate a novel direct relationship between RNA polymerase III transcription and aldolase.
2013 / Revealing properties of the KfrA plasmid protein via combined DLS, AFM and electrokinetic measurements
Adamczyk Zbigniew, Kujda Marta, Nattich-Rak Małgorzata, Marta Ludwiczak, Grażyna Jagura-Burdzy, Małgorzata Adamczyk / Colloids and Surfaces B-Biointerfaces, 2013, vol. 103, s.635-641. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.1016/j.colsurfb.2012.10.065
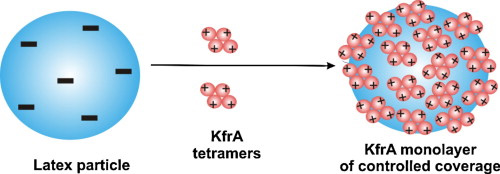
Physicochemical characteristics of the plasmid KfrA protein in electrolyte solutions were done using a combination of dynamic light scattering (DLS), atomic force microscopy (AFM) and electrokinetic methods. The size of the protein was determined via the diffusion coefficient measurements using DLS. It was revealed from these measurements that the protein exists in an aggregated state composed of four molecules. The size of the protein was also precisely determined via AFM imaging of single molecules adsorbed on mica from dilute solutions at pH =3.5. It was 10.6 nm in accordance with the value predicted for an aggregate composed of four monomers in a hexagonal configuration. The aggregation number was also confirmed by kinetics measurements carried out under diffusion controlled transport using AFM imaging of proteins. Further characteristics were acquired via KfrA adsorption on polystyrene latex particles (average size of 820 nm). The electrophoretic mobility of the latex and its zeta potential were determined as a function of the coverage of the protein. The maximum monolayer coverage for pH = 3.5 was 1.2 mg m−2. Additionally, from these measurements the effective charge of KfrA tetramer equal to 12e (elementary charges) was predicted. The KfrA monolayer on latex was used to determine the isoelectric point of the protein, which was pH = 4.5. As concluded, the procedures used in our work proved advantageous for a direct determination of aggregation processes and the effective charge if minor amounts of a protein are available.
2013 / KfrA plasmid protein monolayers on latex particles-electrokinetic measurements
Marta Kujda, Zbigniew Adamczyk, Grazyna Jagura-Burdzy, Małgorzata Adamczyk / Colloids and Surfaces B-Biointerfaces, 2013, vol. 112, s.165-170. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.1016/j.colsurfb.2013.07.026
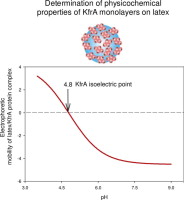
Monolayers of KfrA, a protein assisting in bacteria plasmid segregation, on polystyrene latex particles were produced in controlled self-assembling under diffusion-controlled conditions. The coverage of the protein was quantitatively determined as a function of ionic strength (up to 0.15 M, NaCl) via micro-electrophoretic measurements and concentration depletion with the aid of AFM imaging. The maximum monolayer coverage of KfrA monotonically increased with ionic strength attaining 2.0 mg m−2 for 0.15 M, NaCl that corresponds to the dimensionless coverage of 0.48. This is in accordance with theoretical calculations derived from the random sequential adsorption modeling assuming a tetrameric aggregation state. A high stability of the monolayers in pH cycling experiments was confirmed, which proves the irreversibility of protein adsorption on latex. The acid base properties and the electrokinetic charge of monolayers were also determined via the electrophoretic mobility measurements carried out for various ionic strength. In this way the isoelectric point of the protein of 4.8 was determined, which is prohibitive via bulk measurements. It was concluded that the procedure used in our work is reliable and efficient for characterizing physicochemical properties if minor amounts of a protein are available.
2012 / Engineering of self-sustaining systems: Substituting the yeast glucose transporter plus hexokinase for the Lactococcus lactis phosphotransferase system in a Lactococcus lactis network in silico
Adamczyk Małgorzata, Westerhoff Hans V. / Biotechnology journal, 2012, vol. 7, nr 7, s.877-883. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.1002/biot.201100314
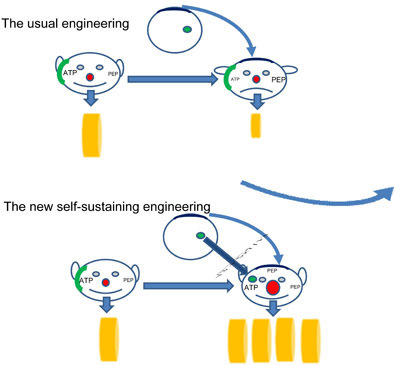
The success rate of introducing new functions into a living species is still rather unsatisfactory. Much of this is due to the very essence of the living state, i.e. its robustness towards perturbations. Living cells are bound to notice that metabolic engineering is being effected, through changes in metabolite concentrations. In this study, we asked whether one could engage in such engineering without changing metabolite concentrations. We have illustrated that, in silico, one can do so in principle. We have done this for the case of substituting the yeast glucose transporter plus hexokinase for the Lactococcus lactis phosphotransferase system, in an L. lactis network, this engineering is ‘silent’ in terms of metabolite concentrations and almost all fluxes.
2011 / Enzyme kinetics for systems biology: when, why and how
Adamczyk Małgorzata, Eunen K., Bakker B., H.V. Westerhoff / Methods in Enzymology, 2011, vol. 500, s.233-257
2010 / Systems biochemistry in practice: experimenting with modelling and understanding regulation and control
H.V. Westerhoff, M. Verma, M. Nardelli, Małgorzata Adamczyk, K. Eunen, E. Simeonidis, B.M. Bakker / Biochemical Society Transactions, 2010, vol. 38, s.1189-1196
2009 / Systems biology: the elements and principles of life
H.V. Westerhoff, C. Winder, H. Messiha, E. Simeonides, Małgorzata Adamczyk, M. Verma, F.J. Bruggeman, W. Dunn / FEBS Letters, 2009, vol. 583, s.3882-3890. Przejdź do dokumentu po identyfikatorze cyfrowym DOI:10.1016/j.febslet.2009.11.018
Systems Biology has a mission that puts it at odds with traditional paradigms of physics and molecular biology, such as the simplicity requested by Occam’s razor and minimum energy/maximal efficiency. By referring to biochemical experiments on control and regulation, and on flux balancing in yeast, we show that these paradigms are inapt. Systems Biology does not quite converge with biology either: Although it certainly requires accurate ‘stamp collecting’, it discovers quantitative laws. Systems Biology is a science of its own, discovering own fundamental principles, some of which we identify here.
2008 / Centromere anatomy in the multidrug resistant pathogen Enterococcus faecium
A. Derome, C. Hoischen, M. Bussiek, R. Grady, Małgorzata Adamczyk, B. Kedzierska, S. Diekmann, D. Barilla, F. Hayes / Proceedings of the National Academy of Sciences of the United States of America, 2008, vol. 105, s.2151-2156
2007 / A zinc-finger like metal binding site in the nucleosome
Adamczyk Małgorzata, Poznański J, Kopera E. , W. Bal / FEBS Letters, 2007, vol. 581, s.1409-1416
2006 / The kfrA gene is the first in a tricistronic operon for survival of IncP-1 plasmid R751
Adamczyk Małgorzata, Dolowy P., Jonczyk M., C.M. Thomas, B. Jagura-Burdzy / Microbiology-Sgm, 2006, vol. 152, nr 6, s.1621-1637
2006 / Współczesne poglądy na kancerogenezę metali- mechanizmy molekularne kancerogenezy jonów niklu
Adamczyk Małgorzata, W. Bal / Bromatologia i Chemia Toksykologiczna, 2006, vol. 39, nr 4, s.361-370
2003 / Erratum: Spread and survival of promiscuous IncP-1 plamsids (Acta Biochimica Polonica (2003) 50:2 (425-453))
Adamczyk Małgorzata, Jagura-Burdzy Grażyna / Acta Biochimica Polonica, 2003, vol. 50, nr 4, s.1283
2003 / Spread and survival of promiscuous IncP-1 plasmids
Adamczyk Małgorzata, Jagura-Burdzy G. / Acta Biochimica Polonica, 2003, vol. 50, s.425-453